 |
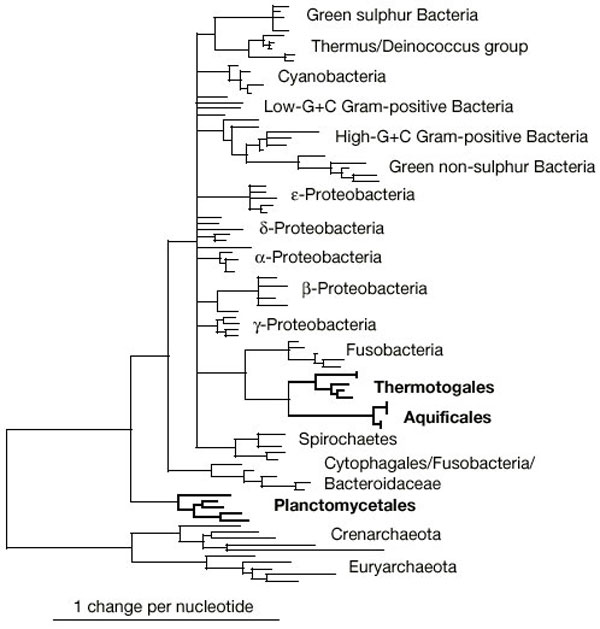
| Figure 1 Prokaryotic phylogenetic tree based on conserved positions in ribosomal RNA. The 'slow–fast' method7 was used to estimate the rate of evolution at each position as the sum of the number of substitutions within 19 predefined phyla. Alignments were constructed for various thresholds (0–15) and the most parsimonious was inferred using PAUP 4b8 (alignments and trees are available from the authors). The relative positions of Planctomycetales, Aquificales and Thermotogales remain very similar for all thresholds from 1 to 10. The phylogeny shown here is based on the 751 positions with a threshold of fewer than 5 substitutions, which represents 45% of informative positions and is thus a valid compromise between the quality and the quantity of phylogenetic information. |